We have been developing software for medical-biological research for many years and can look back on many successfully completed projects, for example in the fields of cell biology, laboratory instrument development, molecular biology, medical research, and bioinformatics.
Project Examples
One of our most recent projects in this field is the 3-year research project that started on 1 February 2021 and is funded by the German Federal Ministry of Education and Research. The research network is composed of five network partners: a research company for applied research, economic companies, and a research institute.
The overall goal of the project is to research a novel platform for digital multiplex assays. The new device should make it possible to analyze a large number of DNA segments (DNA sequences) of a sample particularly quickly, cost-efficiently and in an unprecedented complexity (multiplex degree). In order to determine which DNA segments and in what quantity they are present in the sample, fluorescent molecules - fluorophores - are used to distinguish the DNA. The laboratory instrument will use digital PCR (dPCR) as the basic method.
Polymerase Chain Reaction (PCR)
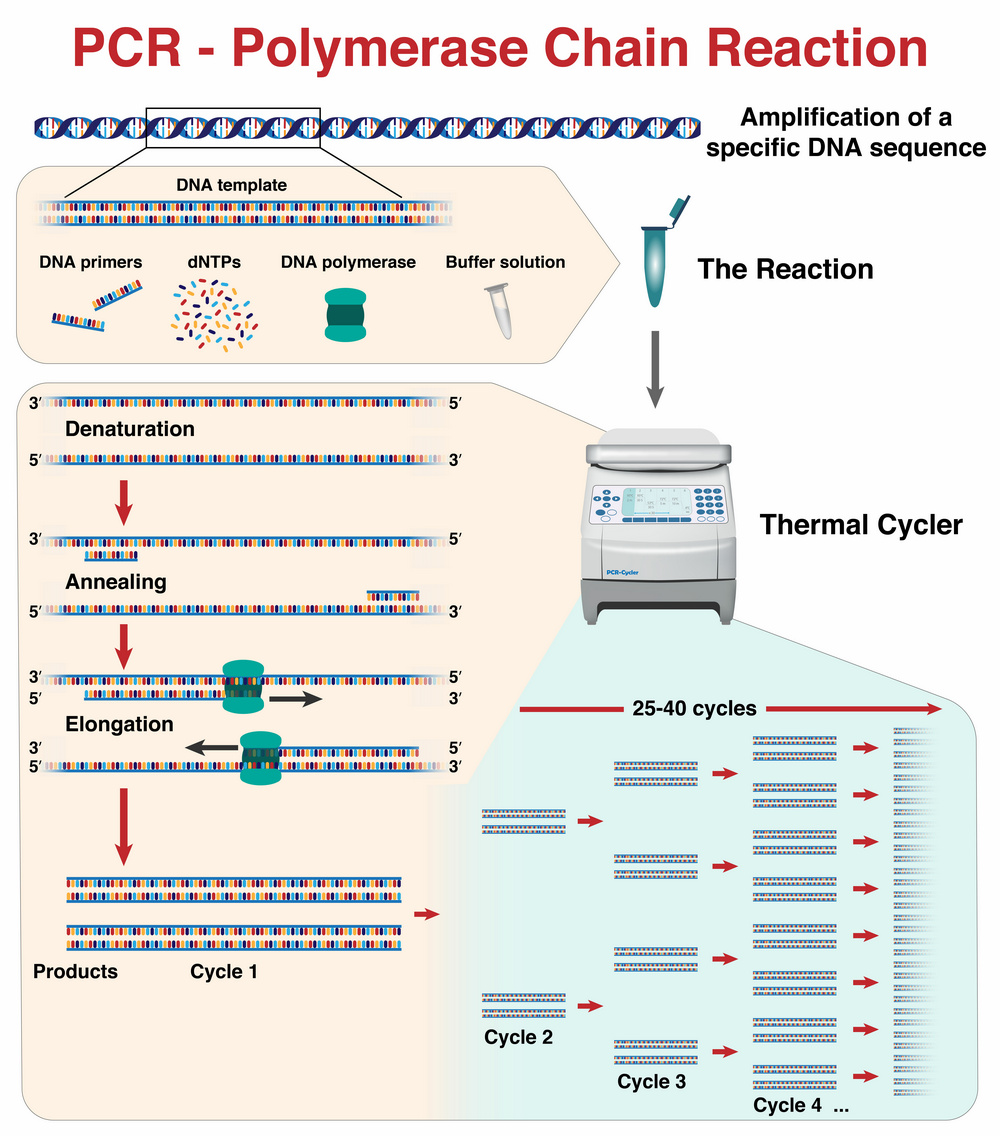
The polymerase chain reaction is a standard method in molecular biology for amplifying DNA. In many successive cycles, the desired DNA segment is amplified in a thermocycler device with the help of the enzyme DNA polymerase. The reaction is called a chain reaction because the DNA that has already been amplified always serves as the starting product for the new exponential amplification cycles (see figure).
Since the invention of PCR by biochemist Karry Mullis in 1983, PCR has been used as one of the most important standard methods in basic molecular biology research as well as in everyday laboratory work. Examples include cloning, the detection of hereditary diseases and the identification of viral infections. It is not without reason that Mullis was awarded the Nobel Prize in Chemistry in 1993 for the development of PCR.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR): Analytical Method for the Detection of COVID-19 Diseases.
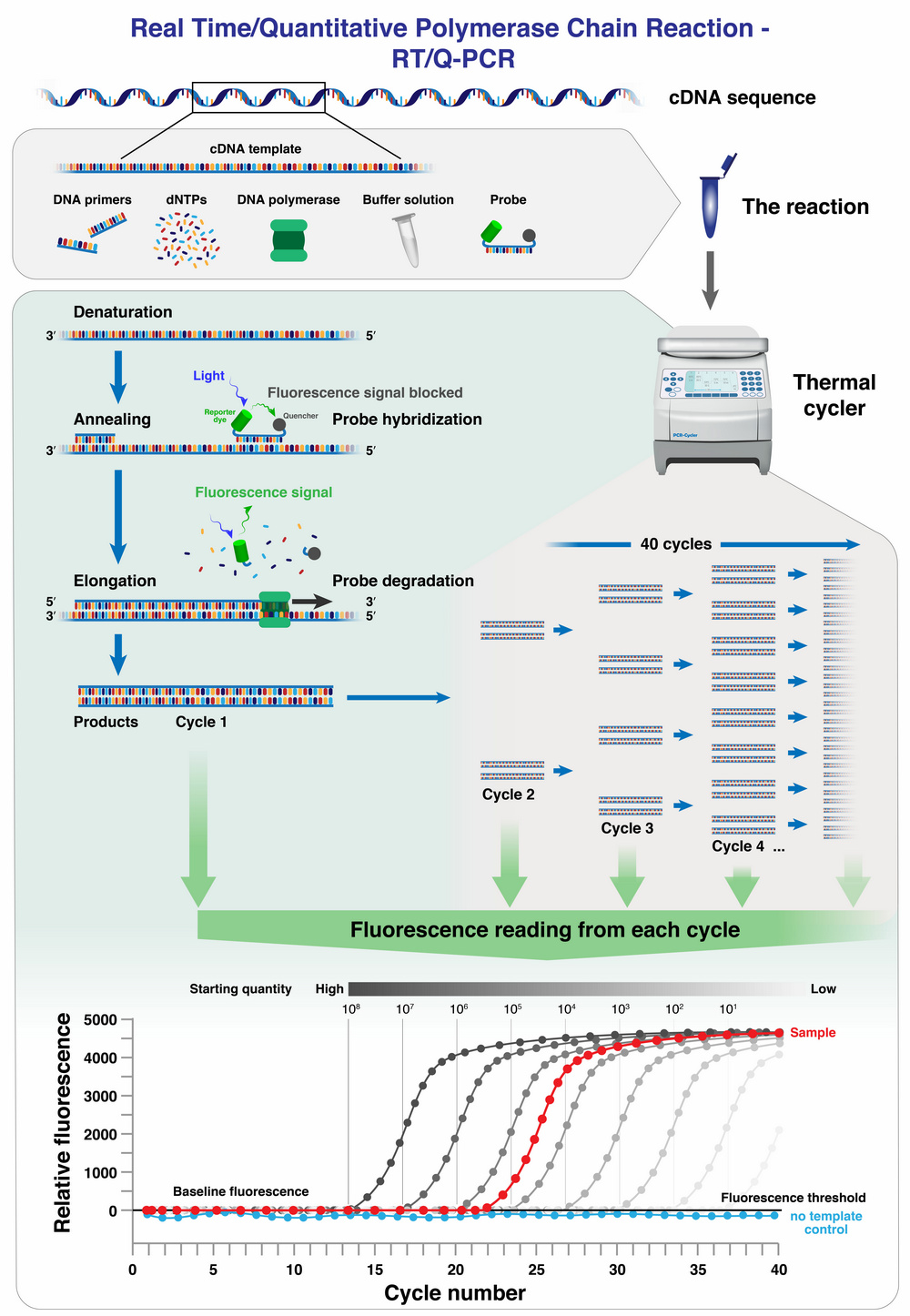
Since the start of the COVID-19 pandemic in 2020, the polymerase chain reaction has become familiar to the world at large. Since the beginning of the pandemic, qRT-PCR (quantitative real-time PCR) has been used to test for SARS-CoV-2. At this time, this is the most accurate and inexpensive method known for analyzing patients for CoV-2 disease.
An extracted DNA sample is placed in a reaction tube along with other reaction solutions and placed in a qRT-PCR instrument. In real-time PCR, fluorescence is measured by the instrument whenever fluorescently labeled DNA probes detect the existence of a particular DNA sequence being sought during the polymerase chain reaction.
Since during a qPCR the target DNA, such as a DNA sequence of SARS-CoV-2, is constantly amplified in a reaction vessel, the intensity of emitted fluorescence also increases over time during the process as the amount of DNA increases.
By comparing the fluorescence intensity of reference samples to the sample under investigation, the presence and quantity of a sought-after DNA sequence in the sample can be determined mathematically after completion of the measurements.
ince this measurement method does not result in a direct measurement of the DNA quantity, using only reference samples, it is not an absolute quantity determination (quantification), but a relative one.
Digitale Polymerase Chain Reaction (dPCR)
Like qRT-PCR, the digital polymerase chain reaction (dPCR) is also used for quantification of a sought DNA sample. Prior to analysis, the reaction mixture is separated into thousands of droplets or compartments and read out individually after analysis. This enables very precise analysis and absolute quantification of DNA sequences. In dPCR, the DNA polymerase also amplifies the DNA under investigation. In this case, however, it does so individually in each reaction droplet or compartment. Whether the DNA sequence of interest is present and in what quantity is determined by the presence of fluorescence.
Since the amplification of the DNA quantity is assumed to be Poisson distributed, i.e. that the amplification of the DNA proceeds uniformly over time, the DNA quantity of the entire starting sample can be determined on the basis of the count of fluorescent and non-fluorescent droplets as well as the total quantity of droplets.
In contrast to qRT-PCR, dPCR is much less sensitive to factors that inhibit the PCR reaction (PCR inhibitors), does not rely on reference samples for quantitation, and can demonstrate a much higher detection limit. Here, a detection limit of a measurement method indicates the extreme value to which a measurand can just be detected. Even though dPCR can provide much more accurate detection of DNA quantities, the major disadvantage so far is that dPCR measurement devices are much more expensive than qRT-PCR devices, which is why the latter are mostly used for measurement. This is exactly where our research project FREEDOM comes in (see above).
Ask us without Obligation
Tell us about your field and let us advise you on existing solutions or individual extensions. Benefit from our many years of experience in image processing.




